Palermo Lab

At PalermoLab, My research focuses on developing and applying of advanced classical and quantum mechanical molecular dynamics (MD) simulations to investigate the conformational dynamics and molecular mechanisms of CRISPR Cas systems.
Additionally, in PalermoLab, we explore the use of artificial intelligence algorithms and frameworks for the advancement of molecular dynamics simulations to explore the rare-event conformational space.
By employing these computational techniques, I aim to gain a fundamental understanding of DNA/RNA molecular systems and gene editors, ultimately providing mechanistic insights that complement and guide rational experimental design.
Sode Lab
Principal Investigator: Dr. Olaseni Sode
Computational Chemistry


Previously, in my masters program at the California State University in Los angeles, I worked in the laboratory of Dr. Olaseni Sode’s laboratory research group. My masters thesis was to characterize weakly attracted van der Waals (vdW) complexes of Rg-CO2 (Rg = He, Ne, Kr, Xe) using quantum chemistry software and machine learning. Understanding these weakly attracted complexes helped to elucidate weak intermolecular interactions of rare-gas systems. This project culminated in my very first first-author publication in 2024: Rodriguez, L.; Natalizio, M.; Sode, O. Theoretical Insights into the Vibrational Structure of Carbon Dioxide Rare-Gas Complexes. J. Phys. Chem. A 2024, 128 (21), 4199–4205. https://doi.org/10.1021/acs.jpca.4c00639
For a brief explanation of the fundamentals and motivation for this research check out my research update from March of 2022.
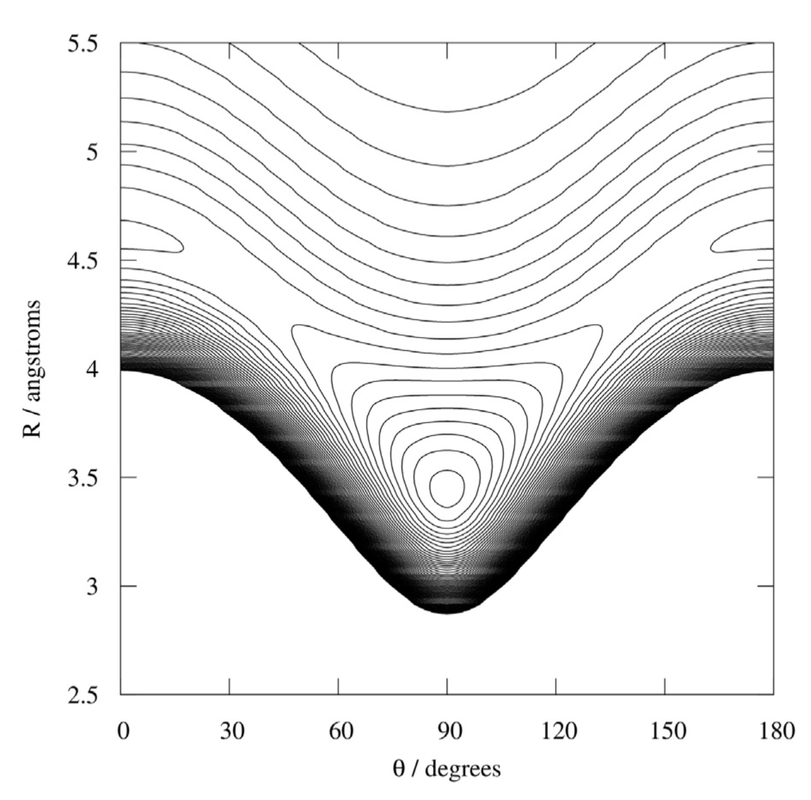
In short, to characterize these weakly interacting complexes we use their vibrational structures. To do this, we constructed a potential energy surface for each of the complexes. This surface is like a piece of paper that is warped in such a way that it has a low point or a divot in its center (an example of this can be found in the research update from March of 2022). The lowest point in the surface is the surface’s global minimum.
In the potential energy surface of Rg-CO$_2$, there are three variables that change the shape of the surface: the intermolecular distance between the carbon dioxide, angle that the carbon dioxide takes relative to the rare-gas (Rg) atom and the energy. When you plot the surface, the intermolecular distance and angle are plotted on the x and y axes and the energy is plotted on the z axis.
The Sode lab has previously published the potential energy surface of the Ar-CO$_2$ complex using a contour plot that I have posted below. On this plot the z-axis is depicted in the contours like a contour map. Recall that the closer the lines are to each other in a contour plot, the steeper the region is on the plot.
Poster Presentations: